Workflow Nodes
Following are the fields required to create each of these nodes:
ETL Job Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'ETL Job' for etl nodes. |
| Resource | Select an etl job from the dropdown list of jobs you have access to. |
| Node Name | Name given to the node for quick and easy identification. |
| Input Configurations | Arguments which can be used in the job. |
ML Model Inference Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'ML Model' for ml model nodes. |
| Resource | Select a machine learning model from the dropdown list of ml models you have access to. |
| Node Name | Name given to the node for quick and easy identification. |
| Input Dataset | The dataset which contains the file to perform ml model inference on. |
| Select Latest File | Selecting yes would automatically pick latest file in the input dataset to perform ml model inference on. |
| File Name Execution Property Key (must be an execution property key) | When the above field is selected as no, you must a file name from the the input dataset. |
| Target Dataset | The dataset to which ml model inference results be written to. |
Email Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'email' for email nodes. |
| Node Name | Name given to the node for quick and easyidentification. |
| Email Recipient (must be an execution property key) | List ofemails addresses to notify to. Eg: ['john.doe@amorphicdata.com','jane.doe@amorphicdata.com']. Note that emails are not delivered to external domains from SES. If needed, please raise a support ticket to bring SES service out of sandbox status. |
| Email Subject (must be an execution property key) | Subject of the email. |
| Email Body (must be an execution property key) | Body of the email. |
Creation of a workflow that predicts readmission risk of patients and notifies them via email:

In the above example, execution properties email_to, email_body are defined as n/a during the workflow creation, during the run time Post_Processing_ETL_Job figures out which email addresses to send emails based on the patient's details returned by risk model and populates email_to, email_body. The code snippets in section workflow execution properties can be used to populate execution properties.
Textract Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Textract' for textract nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Input Dataset | The dataset which contains files to extract text from. Supported files are of type - PDF, JPG, PNG. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Features | Choose one or more features that you want to extract to target dataset. Text, Forms and Tables are three features available. Text: extracts all text from document Forms: extracts forms as key-value pairs Tables: extracts tables in csv format |
| Target Dataset | The dataset to which extracted text is written to. |
The picture below shows a textract node being created with source dataset - patient_cioms_docs_pdf and target dataset
- patient_cioms_text with file processing mode: all and text as the feature to be extracted to output.

List of files in the source dataset - cioms_john_doe.pdf, cioms_jane_doe.pdf.

List of files in the target dataset post workflow execution
- cioms_john_doe.pdf-text.txt, cioms_jane_doe.pdf-text.txt.

Textract node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.

No two textract nodes can have same input dataset in a workflow.
Rekognition Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Rekognition' for rekognition nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Input Dataset | The dataset which contains files to extract text from. Supported files are of type - MP4, JPG, PNG. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Features | Choose one or more features that you want to extract to target dataset. Text, Faces, Content Moderation, Celebrities and Labels are five features available. Text: extracts all text from document Faces: detects faces from image or video Content Moderation: extracts the inappropriate, unwanted, or offensive content analysis results Celebrities: extracts the name and additional information about a celebrity Labels: extracts label name, the percentage confidence in the accuracy of the detected label |
| Target Dataset | The dataset to which extracted json is written to. |
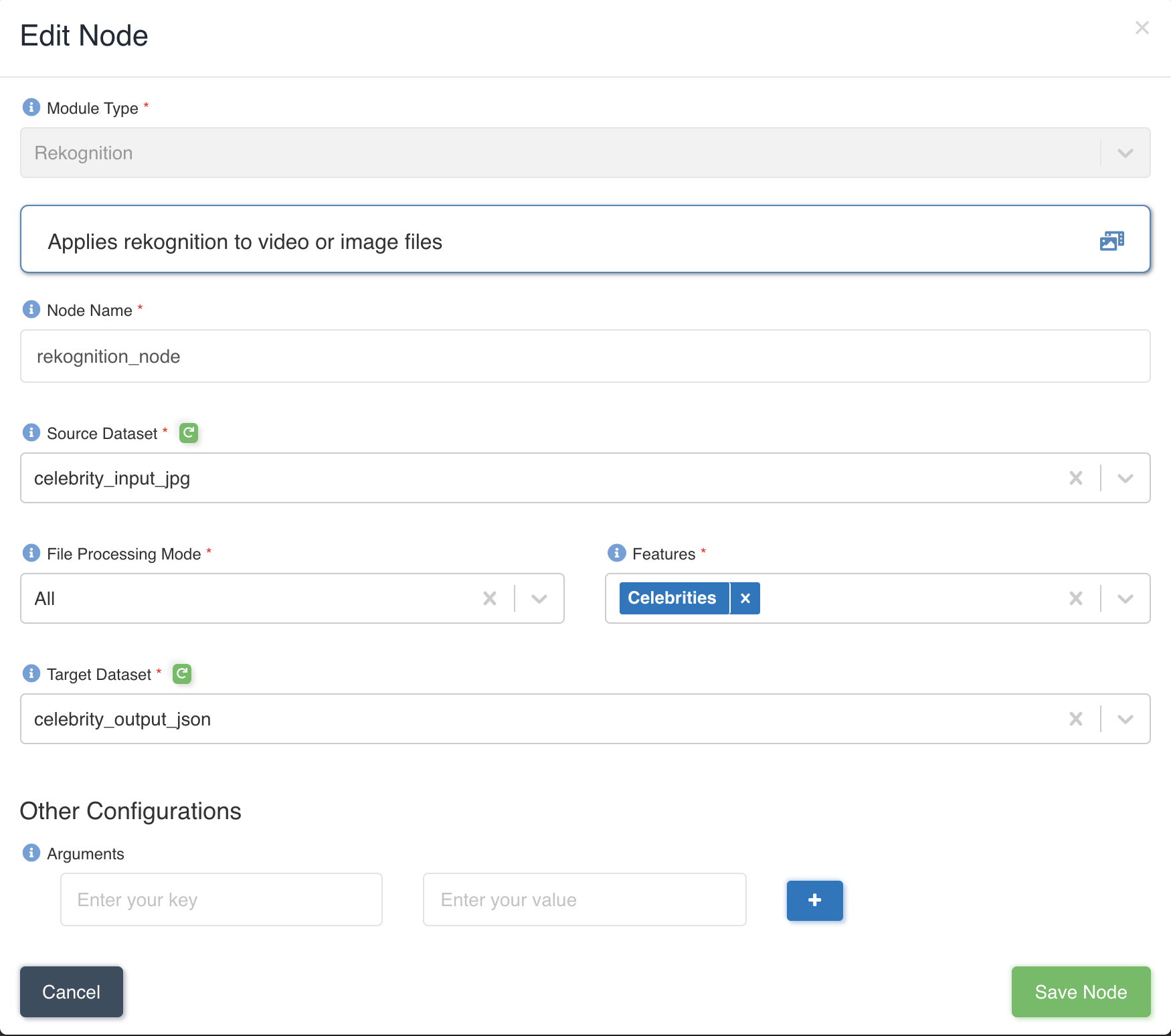
The picture below shows a rekognition node being created with source dataset - celebrity_input_jpg and target dataset
- celebrity_output_json with file processing mode: all and Celebrities as the feature to be extracted to output.
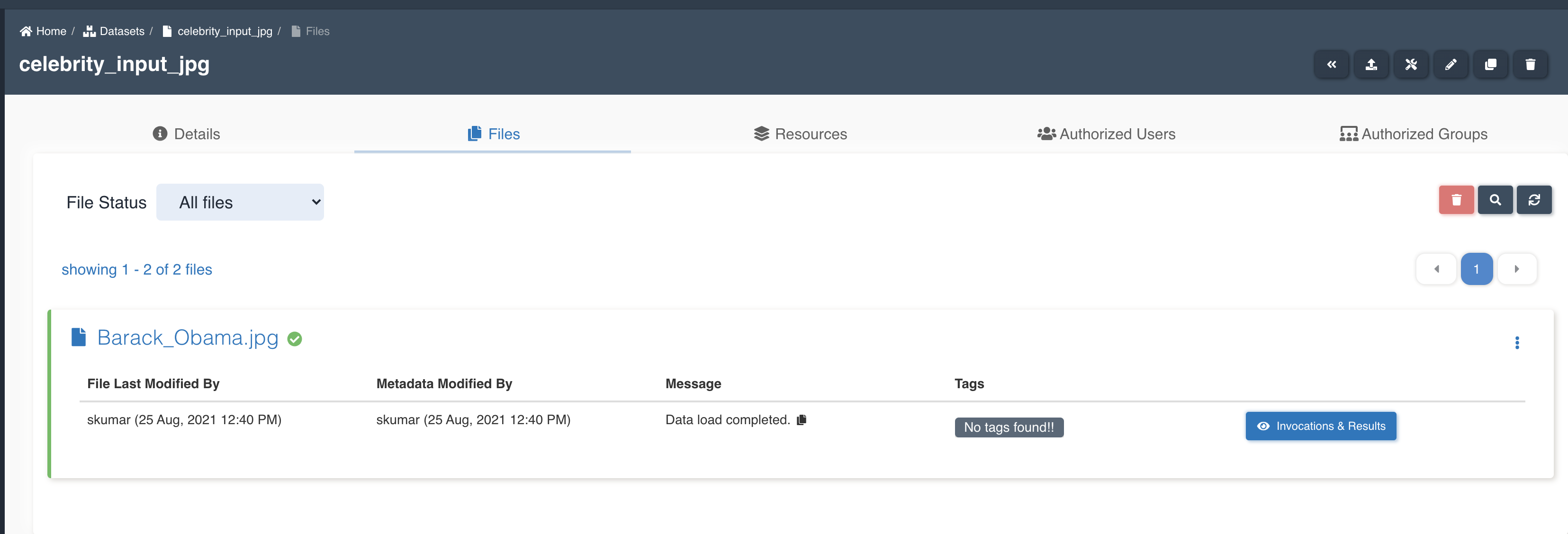
List of files in the source dataset - celebrity_input_jpg.
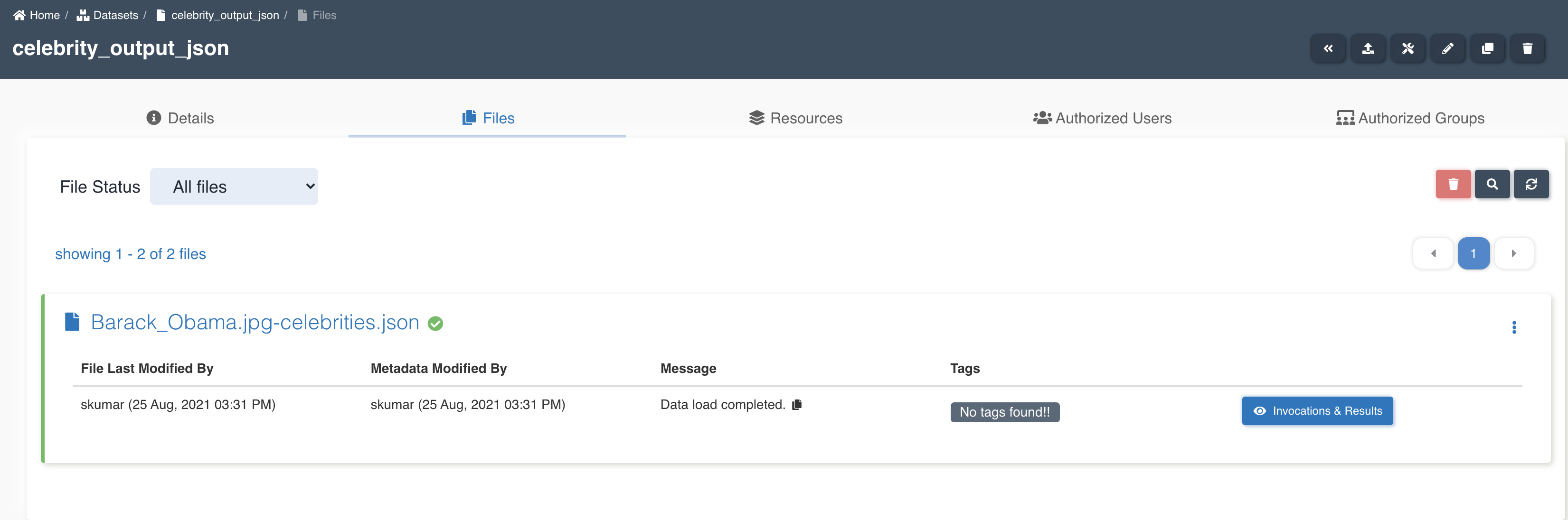
List of files in the target dataset post workflow execution
- barack_obama.jpg-celebrities.json
Rekognition node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.
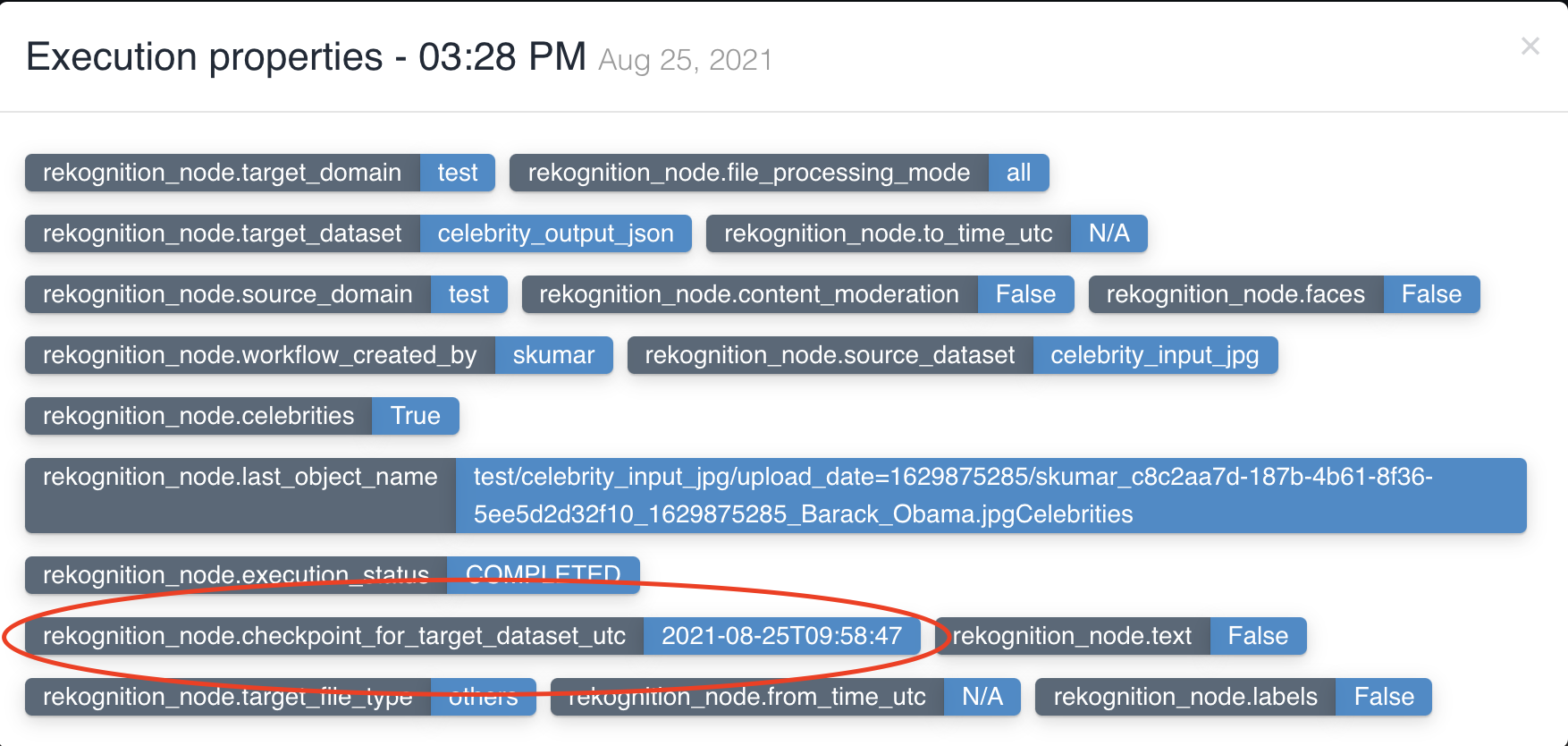
No two rekognition nodes can have same input dataset in a workflow.
Translate Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Translate' for translate nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Source Dataset | The dataset which contains files to translate text from. Supported files are of type - TXT. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Source Language | The language of the text to be translated in source dataset. |
| Target Language | The language to which text is translated to and written to target dataset. |
| Target Dataset | The dataset to which to translate text is written to. Supported files are of type - TXT. |
The picture below shows a translate node being created with source dataset - german_news_articles and target dataset
- german_news_articles_translated_into_english with file processing mode: all.

List of files in the source dataset - news_german_nov_04.text, news_german_nov_03.text, news_german_nov_02.text

List of files in the target dataset post workflow execution - news_german_nov_04.text, news_german_nov_03.text, news_german_nov_02.text.

Translate node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.

No two translate nodes can have same input dataset in a workflow.
Limits: Currently translate node only translates the first 5000 characters of the text, rest of the text is ignored.
Comprehend Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Comprehend' for comprehend nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Input Dataset | The dataset which contains files to find insights and relationships in text. Supported files are of type - TXT. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Features | Choose one or more features that you want to extract to target dataset. Entities, KeyPhrases and Sentiment are three features available. Entities: named entities like people, places, locations etc., in a document KeyPhrases: key phrases or talking points in a document Sentiment: overall sentiment of a text (positive, negative, neutral or mixed) |
| Target Dataset | The dataset to which insights and relationships in text is written to. |
The picture below shows a comprehend node being created with source dataset - sample_news_articles and target dataset
- news_articles_extracted_entities with file processing mode: all and three features - Entities, KeyPhrases, Sentiment to be extracted to output.

List of files in the source dataset - sample_news_article.txt.

List of files in the target dataset post workflow execution - sample_news_article.txt-entities.json, sample_news_article.txt-keyphrases.json.

Comprehend node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.

No two comprehend nodes can have same input dataset in a workflow.
Medical Comprehend Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Medical Comprehend' for medical comprehend nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Input Dataset | The dataset which contains files to find medications, medical conditions etc., in text. Supported files are of type - TXT. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Features | Choose one or more features that you want to extract to target dataset. Medications, Medical conditions, Personal health information and Medical entities are the features available. Medications: Detects medication and dosage information for the patient. Medical conditions: Detects the signs, symptoms, and diagnosis of medical conditions. Personal health information: Detects the patient's personal information. Medical entities: All the medical and personal information in the document |
| Target Dataset | The dataset to which extracted medical information in text is written to. |
The picture below shows a medical comprehend node being created with source dataset - medical_articles and target dataset - medical_articles_extracted_information with file processing mode: all and features - Medications, MedicalConditions, PersonalHealthInformation, MedicalEntities to be extracted to output.

List of files in the source dataset - cancer_medication_article.txt.

List of files in the target dataset post workflow execution - cancer_medication_article.txt-medications.json, cancer_medication_article.txt-medicalconditions.json.

Medical comprehend node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.

No two medical comprehend nodes can have same input dataset in a workflow.
Transcribe Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Transcribe' for transcribe nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Source Dataset | The dataset which contains audio files to extract content in the form of text. Supported files are of type - MP3, WAV. |
| Source Language | The language of the audio files in input dataset. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Features | Choose one or more features that you want to extract to target dataset. Text, ConversationBySpeaker, RedactedText and RedactedConversationBySpeaker are four features available. Text: Raw text extracted from the audio file. ConversationBySpeaker: Raw conversation displaying speaker and the sentence the speaker spoke. RedactedText: Extracted text from audio file with some content obscured for legal and security purposes. RedactedConversationBySpeaker: Conversation displaying speaker and the sentence the speaker spoke with some content obscured for legal and security purposes. |
| Target Dataset | The dataset to which extracted content is written in the form of txt files. |
The picture below shows a transcribe node being created with source dataset - sample_news_articles_mp3 and target dataset - sample_news_articles_extracted_entities with file processing mode: all and four features - Text, ConversationBySpeaker, RedactedText and RedactedConversationBySpeaker to be extracted to output.
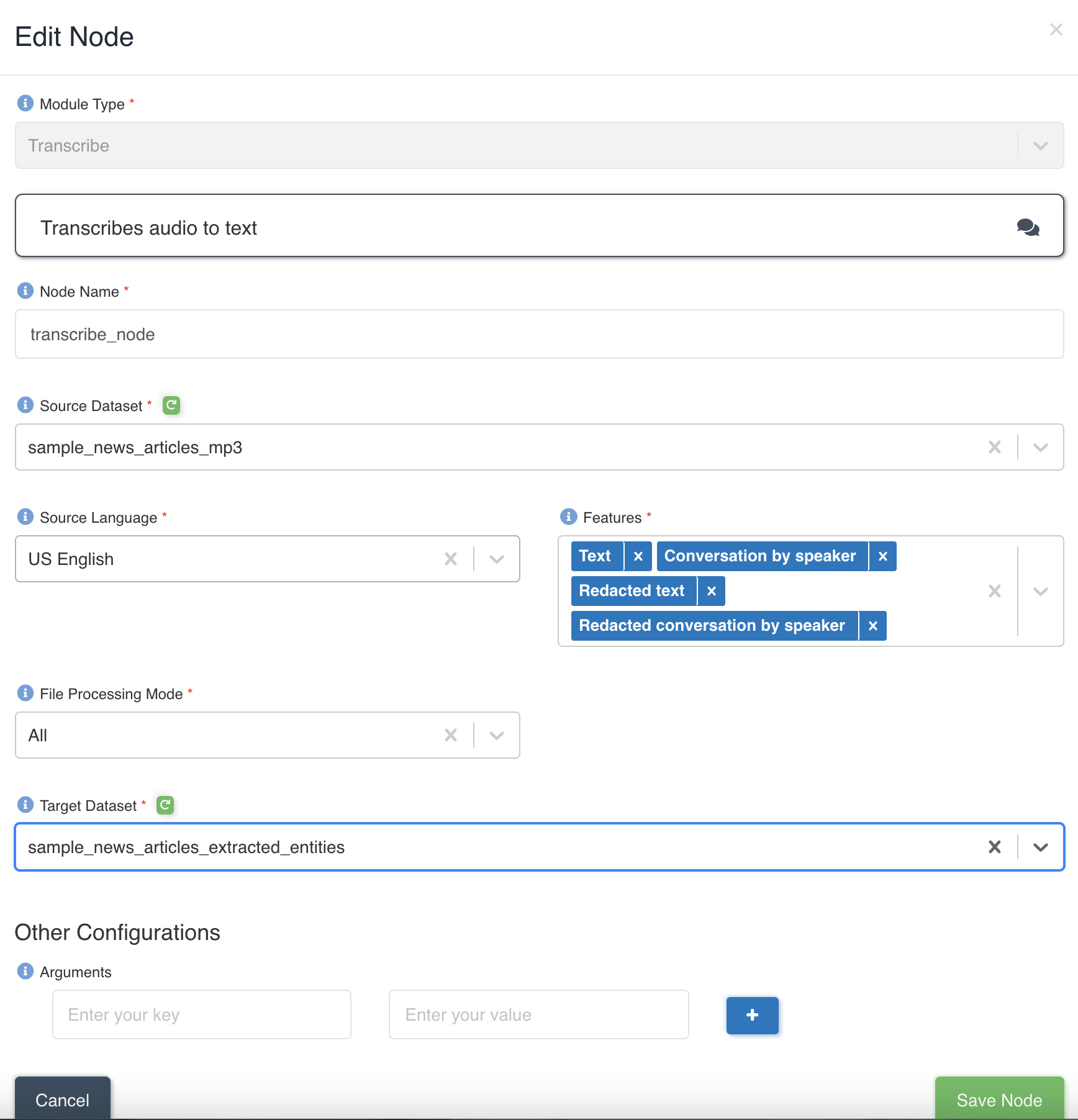
List of files in the source dataset - sample_news_article.mp3.
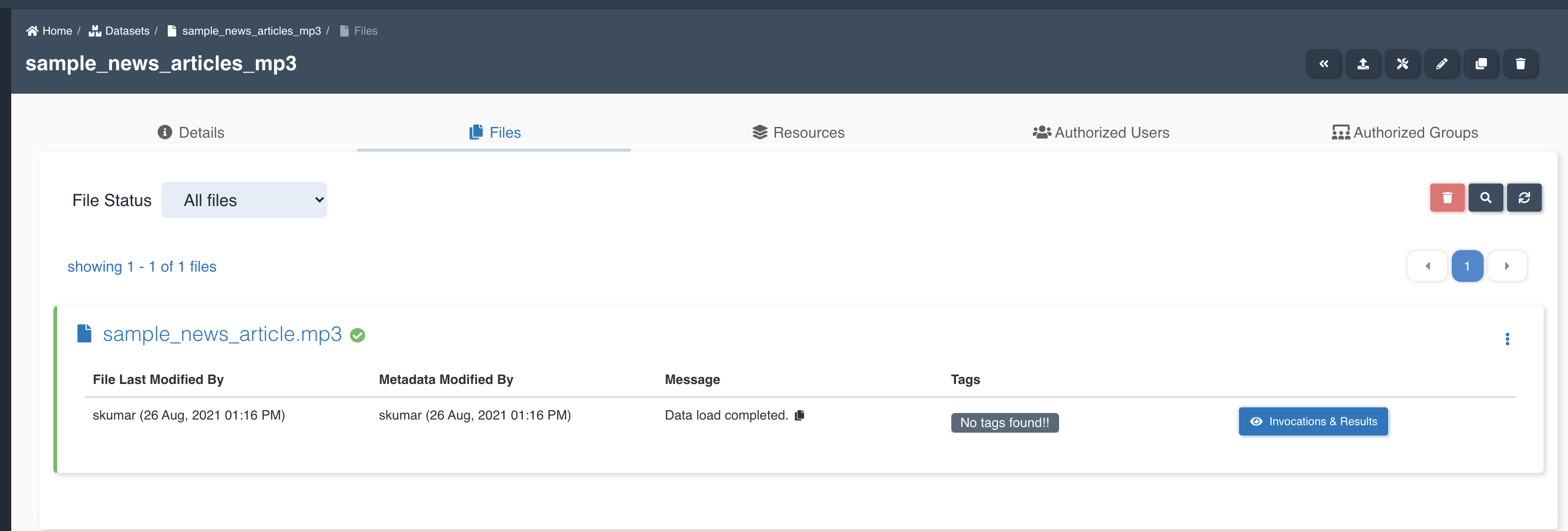
List of files in the target dataset post workflow execution - sample_news_article.mp3-redacted_conversation_by_speaker.txt, sample_news_article.mp3-redacted_text.txt, sample_news_article.mp3-conversation_by_speaker.txt, sample_news_article.mp3-text.txt.
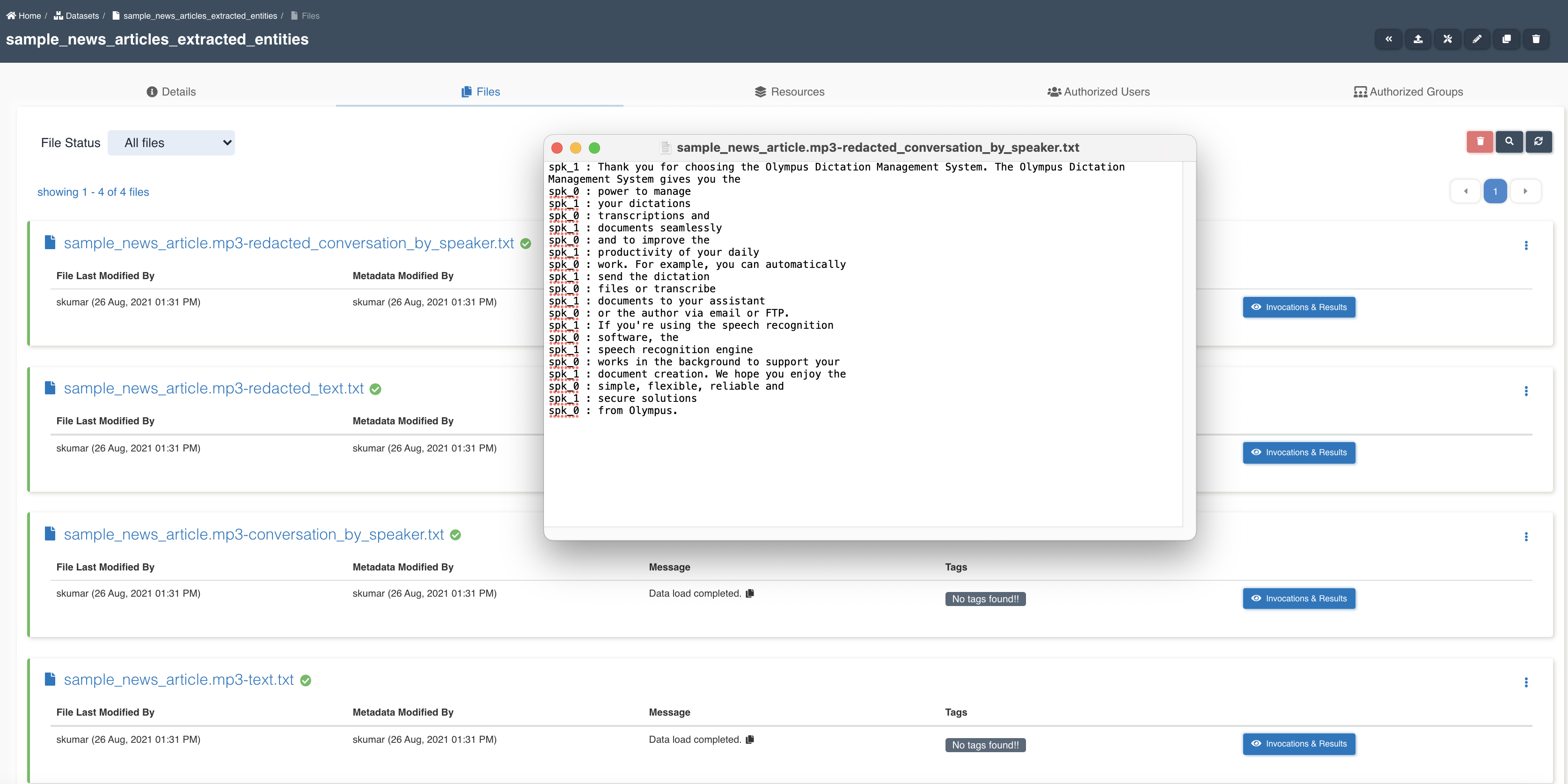
transcribe node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.
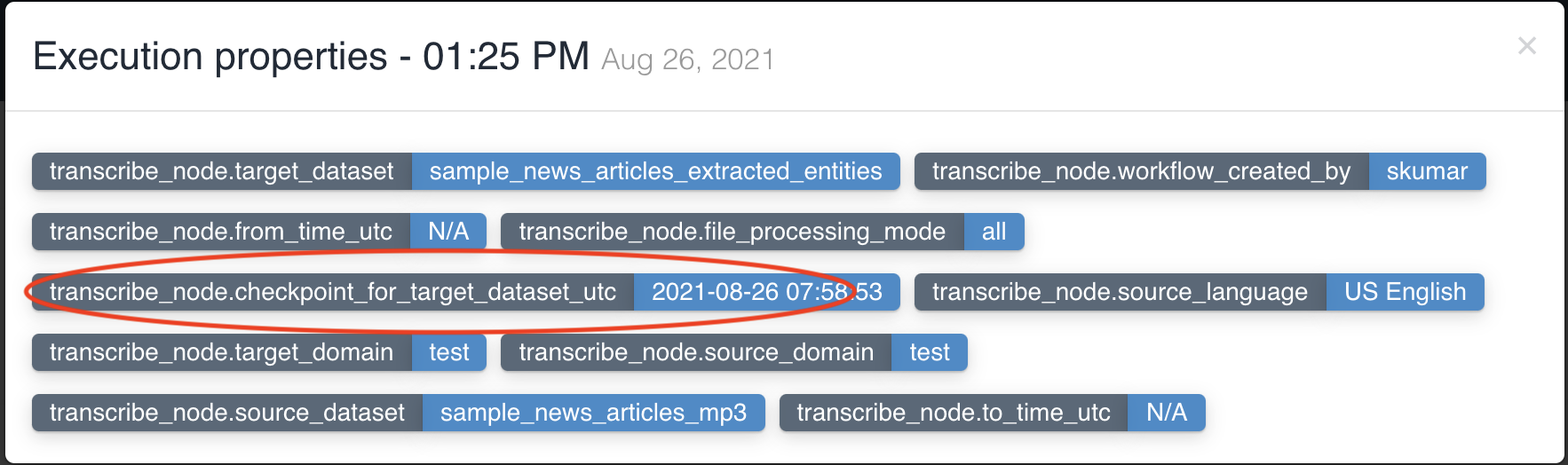
No two transcribe nodes can have same input dataset in a workflow.
Medical Transcribe Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Medical Transcribe' for medical transcribe nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Source Dataset | The dataset which contains audio files to extract content in the form of text. Supported files are of type - MP3, WAV. |
| Source Language | The language of the audio files in input dataset only English-US (en-US) is supported as of now. |
| File Processing Mode | All, Change Data Capture and Time Based are three modes available. All: processes all documents in a source dataset Change Data Capture: processes the documents that have landed after the previous workflow execution Time Based: processes the documents based on the custom time period chosen |
| Features | Choose one or more features that you want to extract to target dataset. Text and ConversationBySpeaker are two features available. Text: Raw text extracted from the audio file. ConversationBySpeaker: Raw conversation displaying speaker and the sentence the speaker spoke. |
| Target Dataset | The dataset to which extracted content is written in the form of txt files. |
The picture below shows a medical transcribe node being created with source dataset - sample_medical_article_mp3 and target dataset - sample_medical_article_extracted_entities with file processing mode: all and four features - Text and ConversationBySpeaker to be extracted to output.
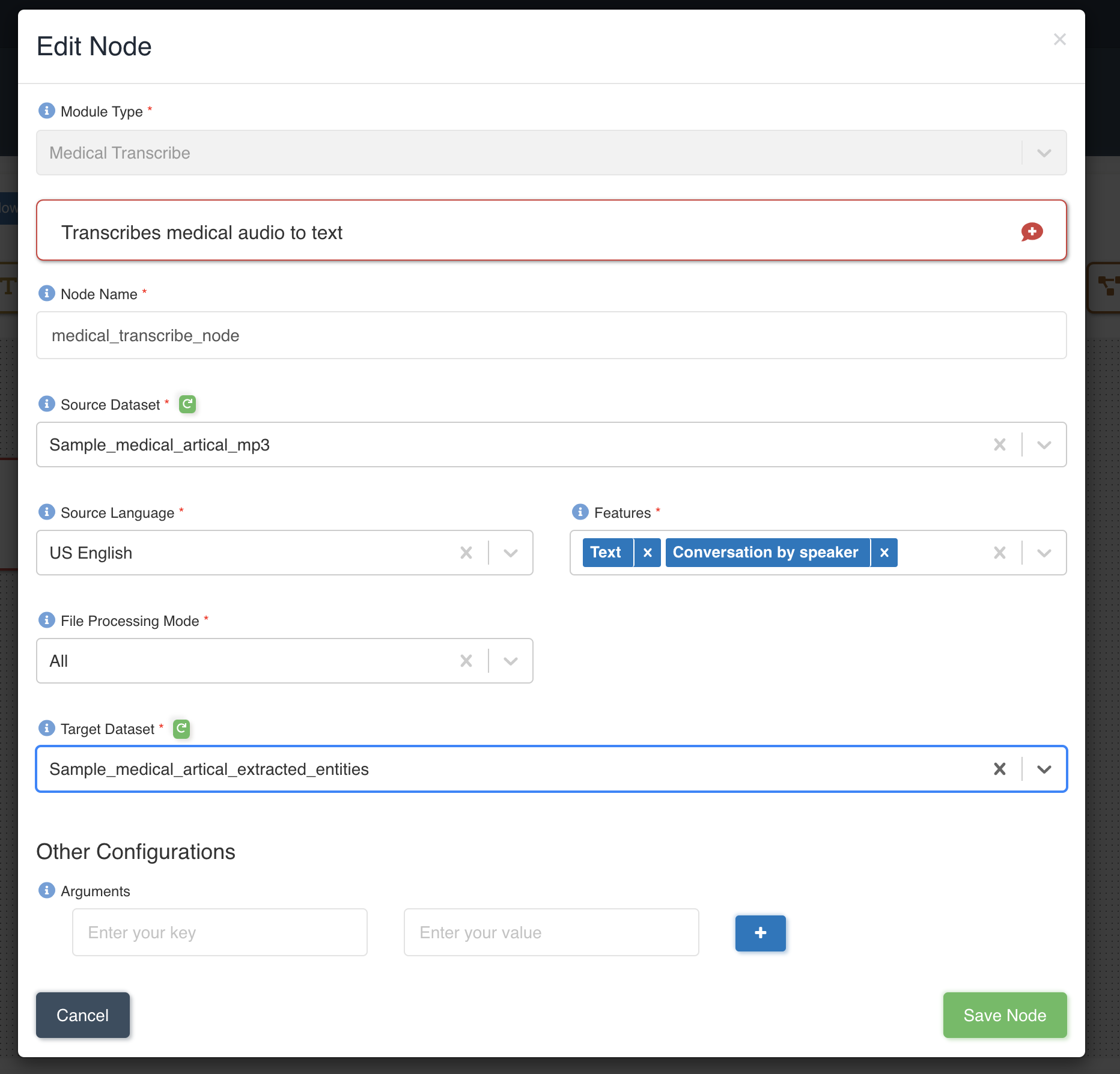
List of files in the source dataset - sample_medical_article.mp3.
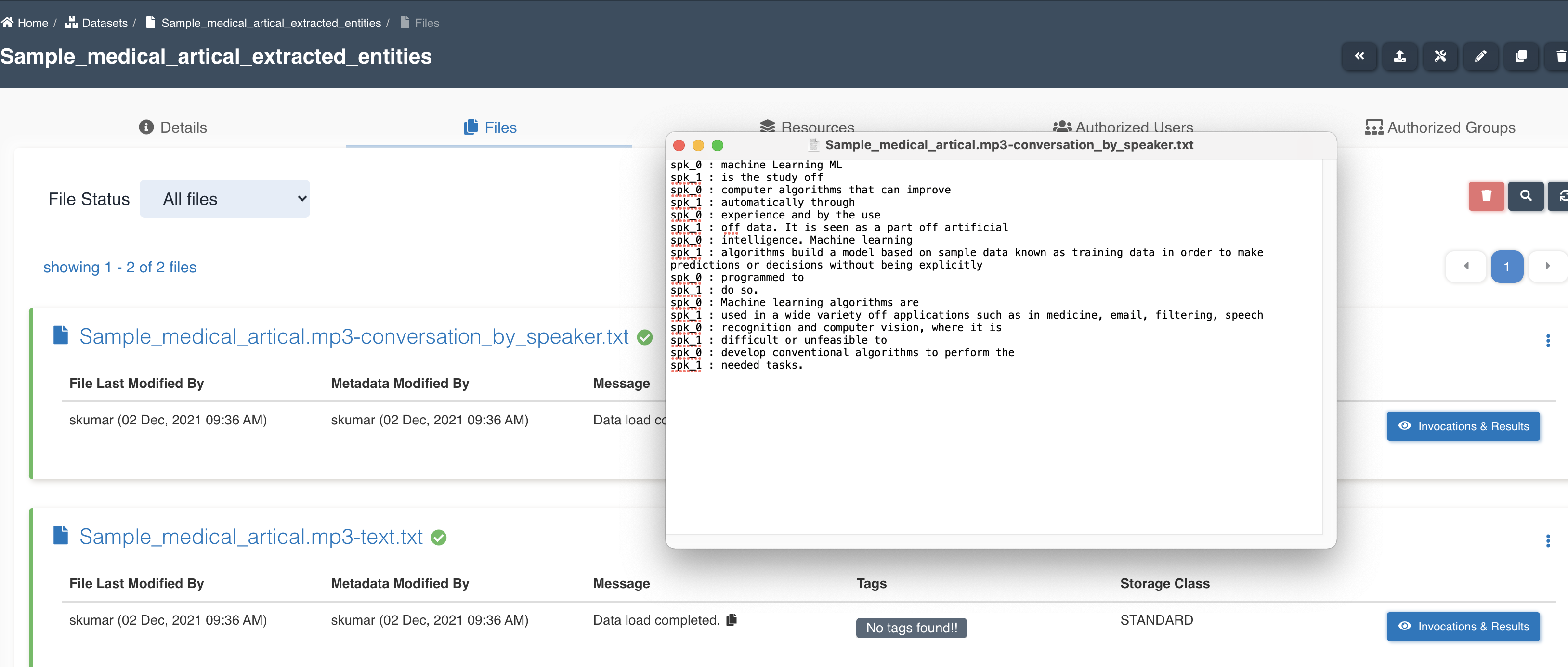
List of files in the target dataset post workflow execution - sample_medical_article.mp3-conversation_by_speaker.txt, sample_medical_article.mp3-text.txt.
medical transcribe node updates workflow execution properties with checkpoint_for_target_dataset_utc so that subsequent nodes can identify files generated from previous node in workflow execution.
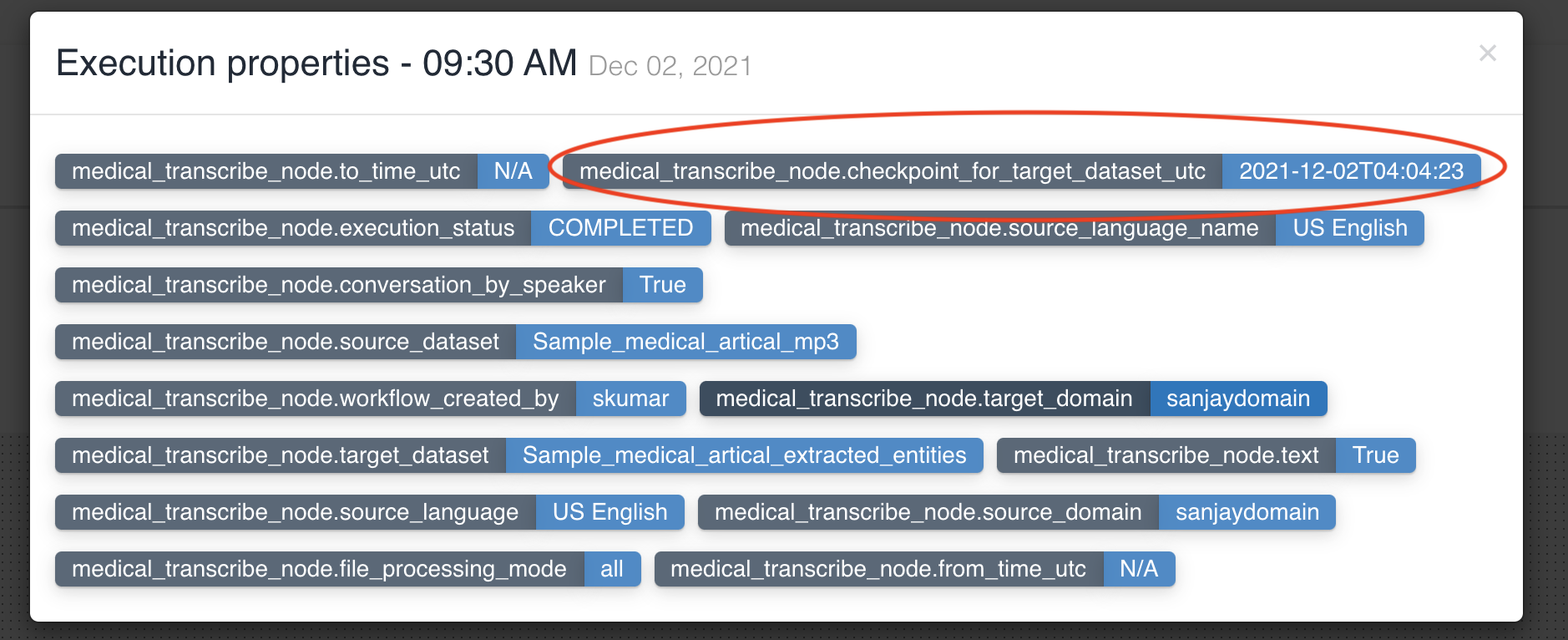
No two medical transcribe nodes can have same input dataset in a workflow.
Workflow Node
Users can create a dependency graph with multiple workflows to solve complex use cases.
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Workflow' for workflow nodes. |
| Resource | Select a workflow from the dropdown list of workflows you have access to. |
| Node Name | Name given to the node for quick and easy identification. |
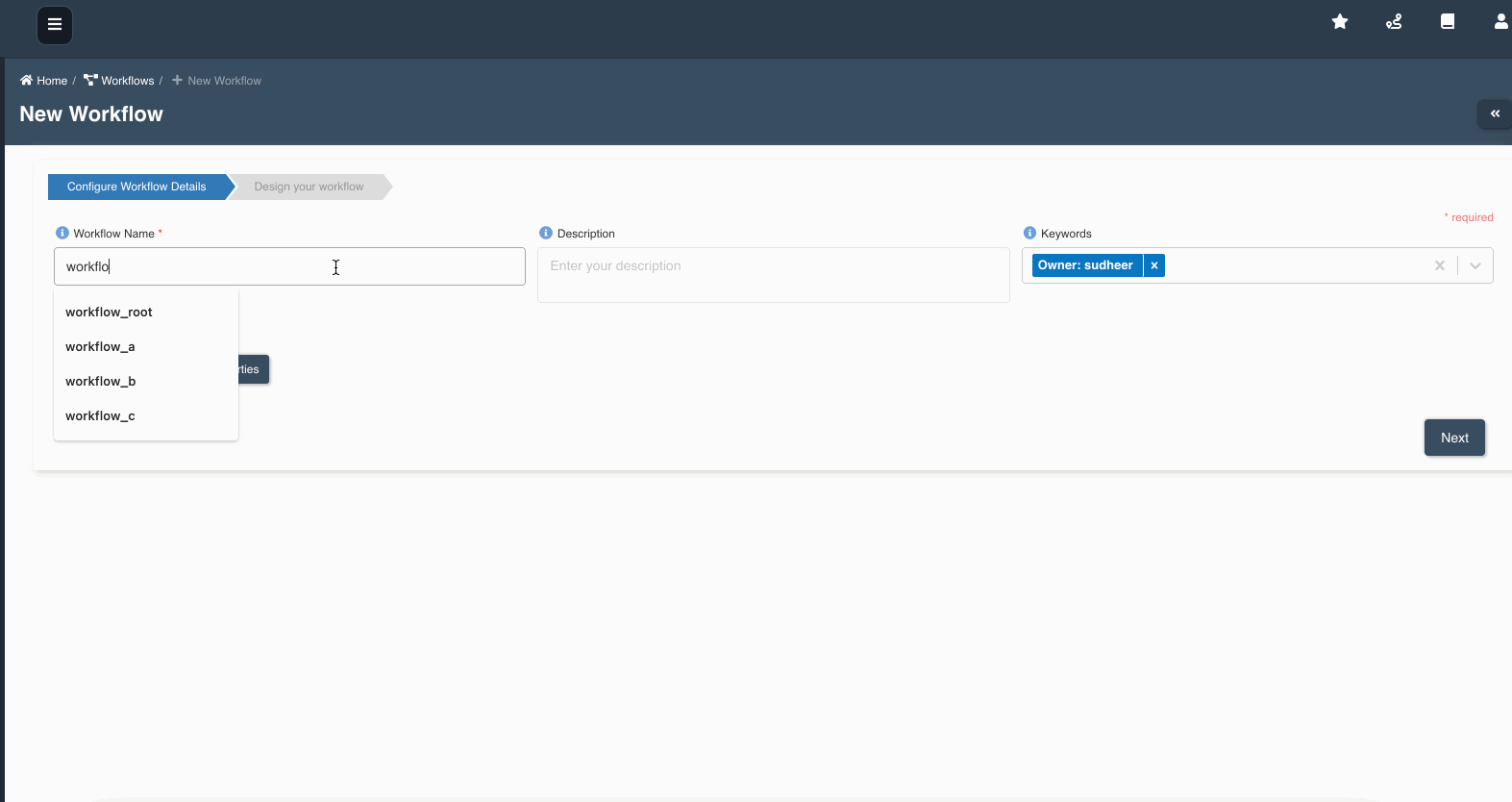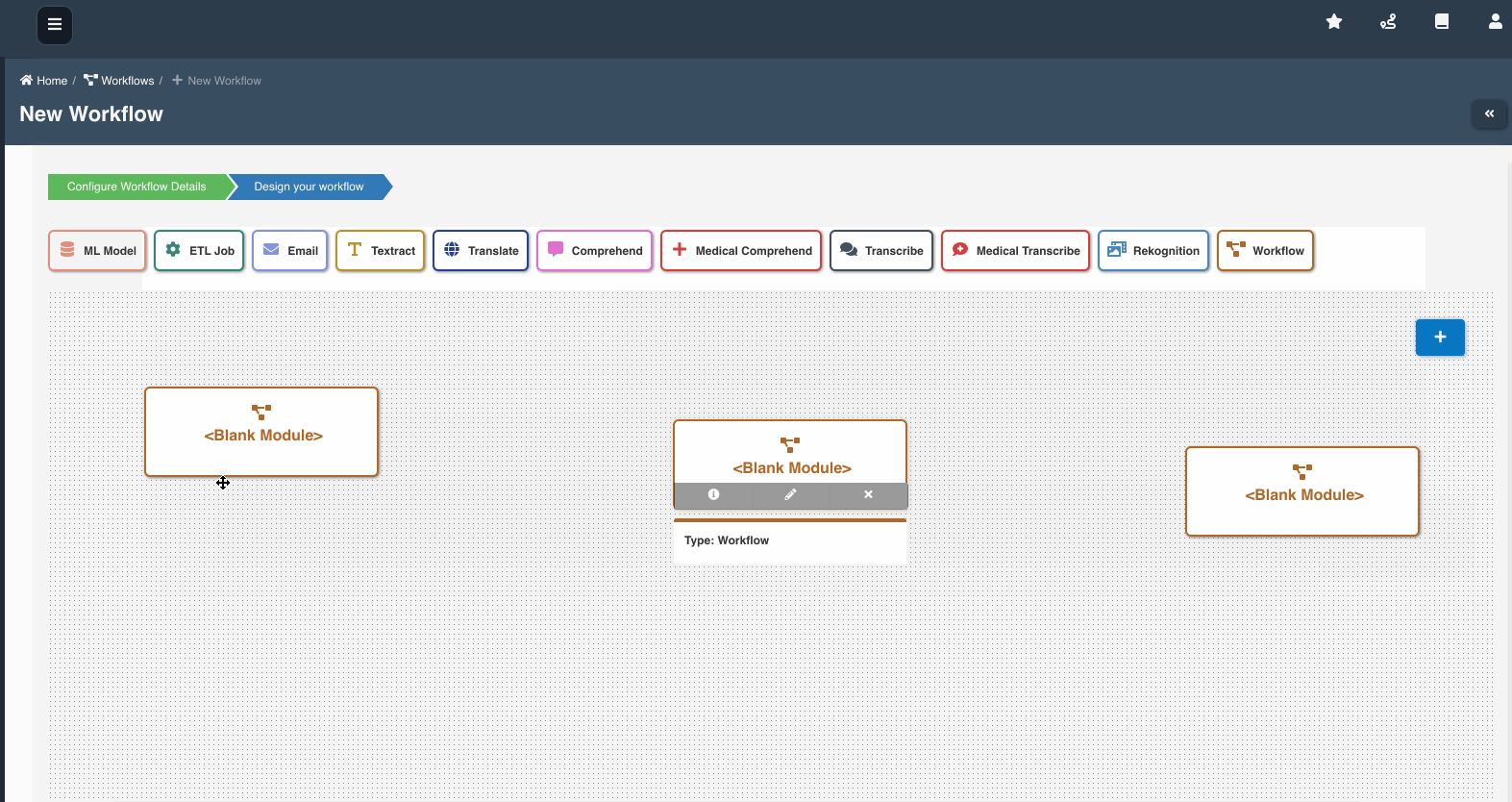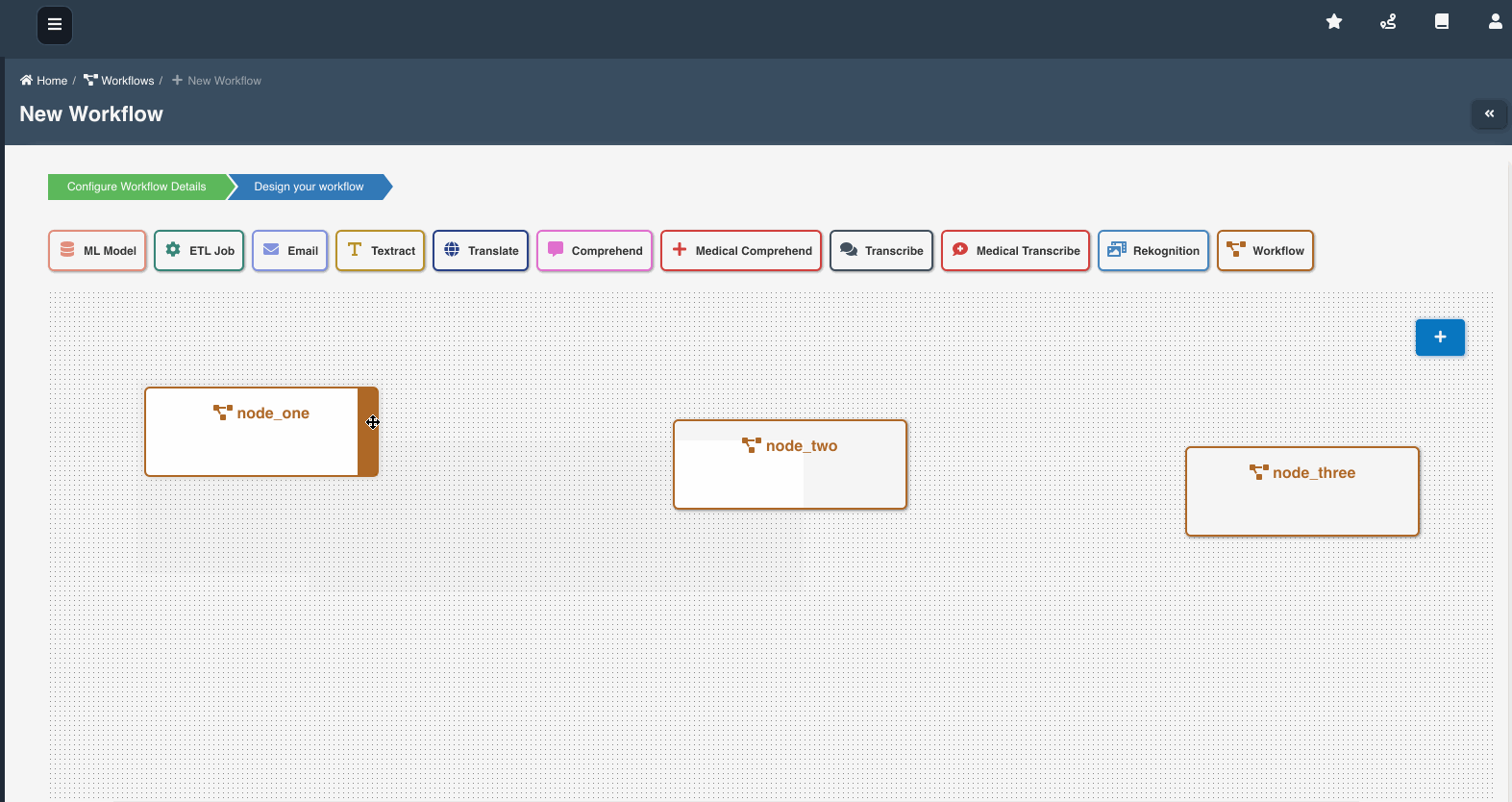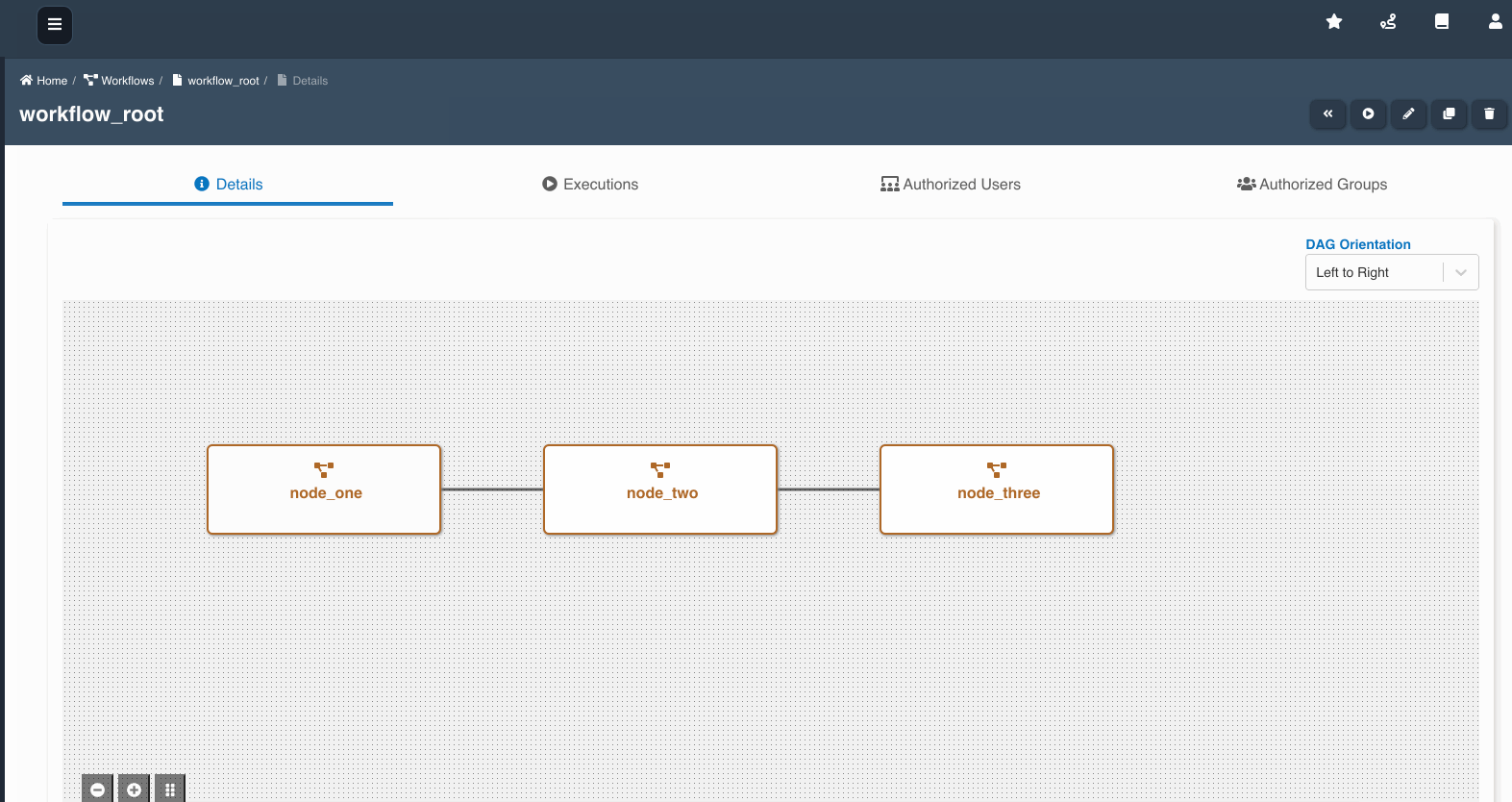
If multiple workflows workflow_a, workflow_b, workflow_c are chained as nodes in workflow_root, we refer workflow_a, workflow_b, workflow_c as child workflows and workflow_root as parent workflow.
Once a parent workflow is stopped, it triggers the stoppage of all of its child workflows.
The following graphic shows how a workflow with workflow nodes is created:
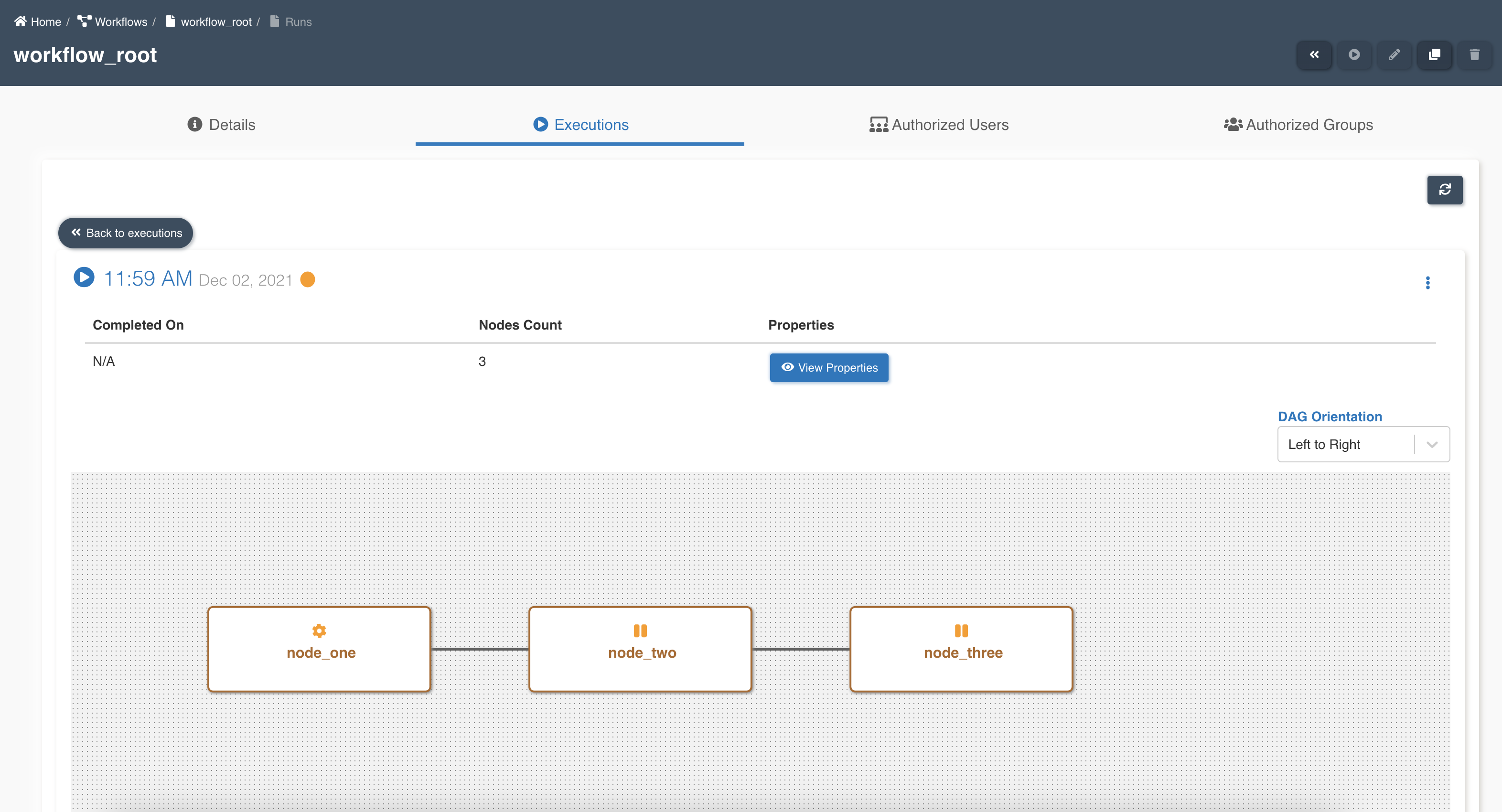
Parent workflow execution shows details of node execution status:
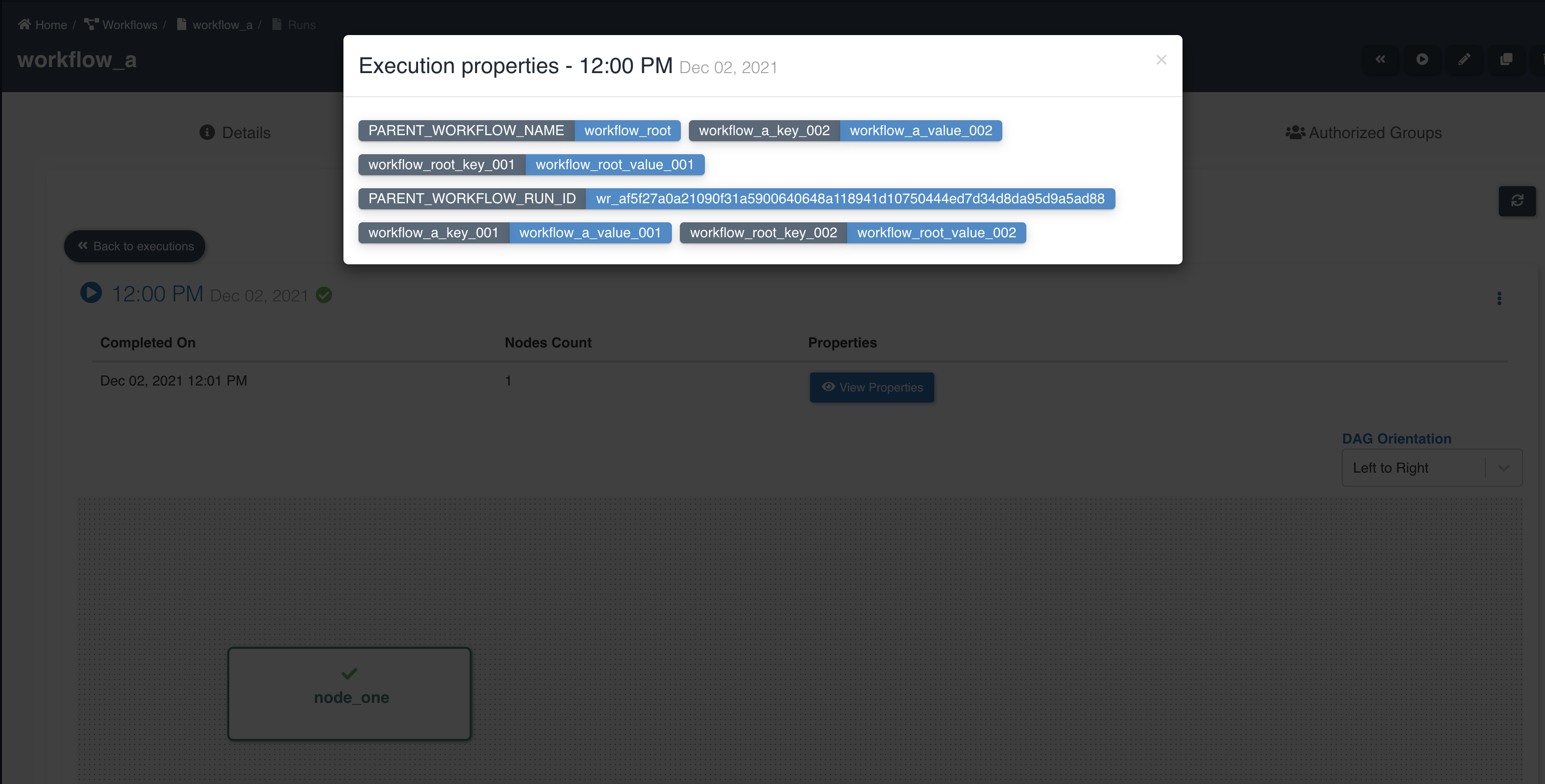
Workflow execution properties can be defined at parent workflow level and they can be accessed and updated from any of the child workflows. Execution properties of a parent workflow overrides execution properties of child workflow. Refer to workflow execution properties for creating execution properties.
Child workflow execution shows details of node execution status and properties inherited from parent workflow:
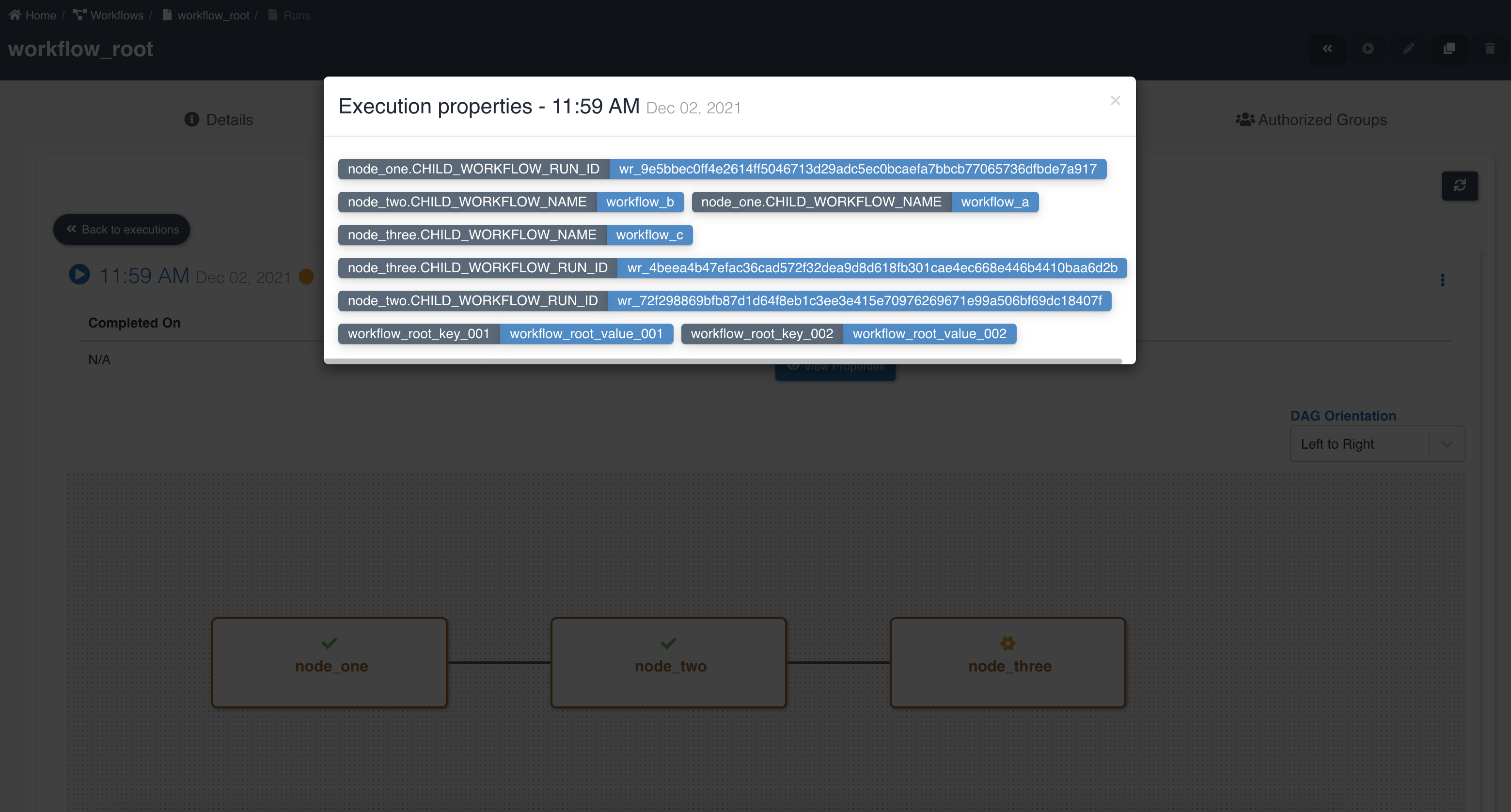
Below graphic showing parent workflow execution properties and details of child workflows:
File Load Validation Node
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'File Load Validation' for file load validation nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Timeout | Optional field to set the timeout value (in minutes) for the node to stop execution. Default is set to 60 minutes. |
The following graphic shows how a file load validation node can be created to validate if all files written by an etl job to a dataset are ready to be consumed:
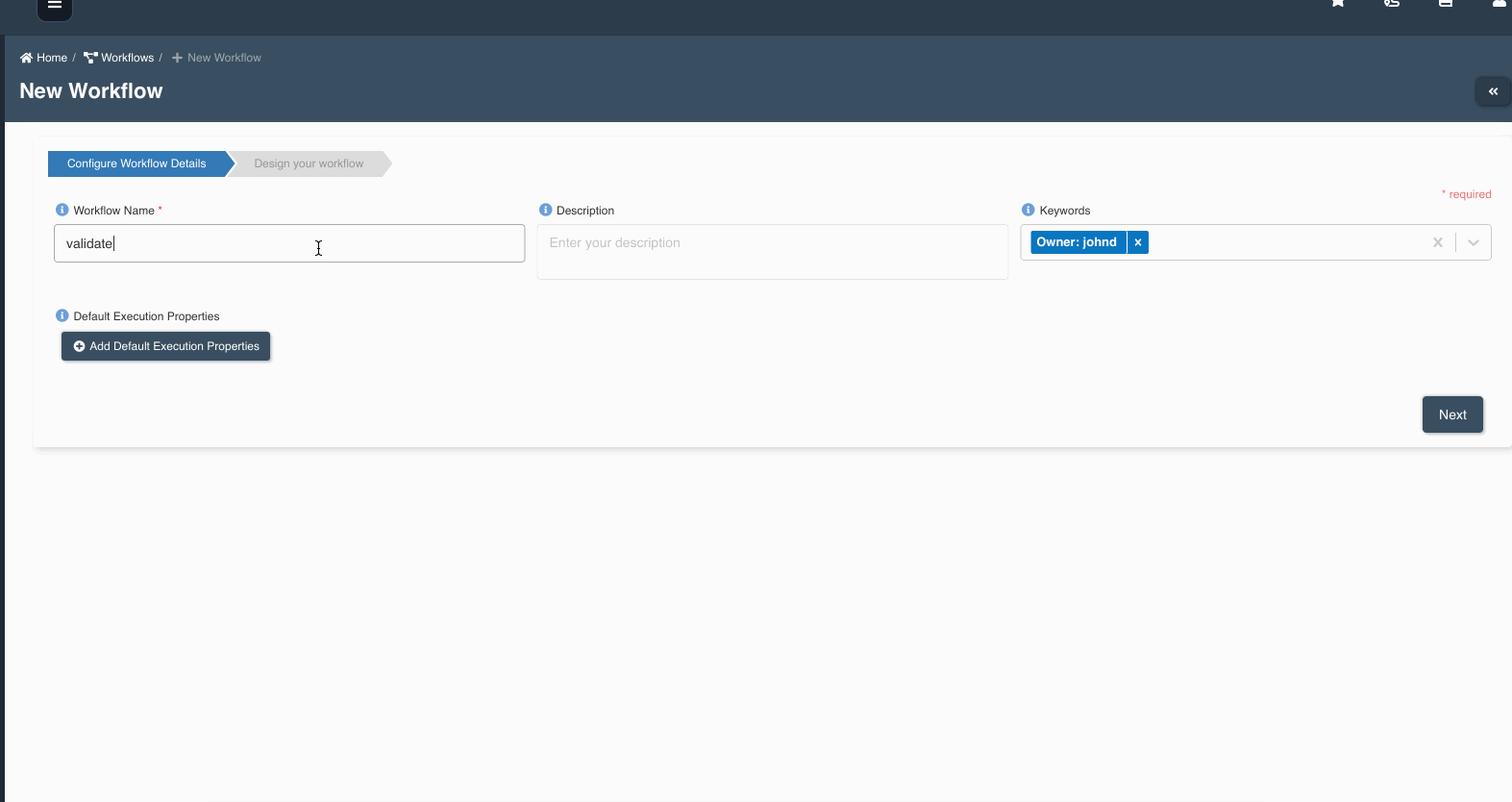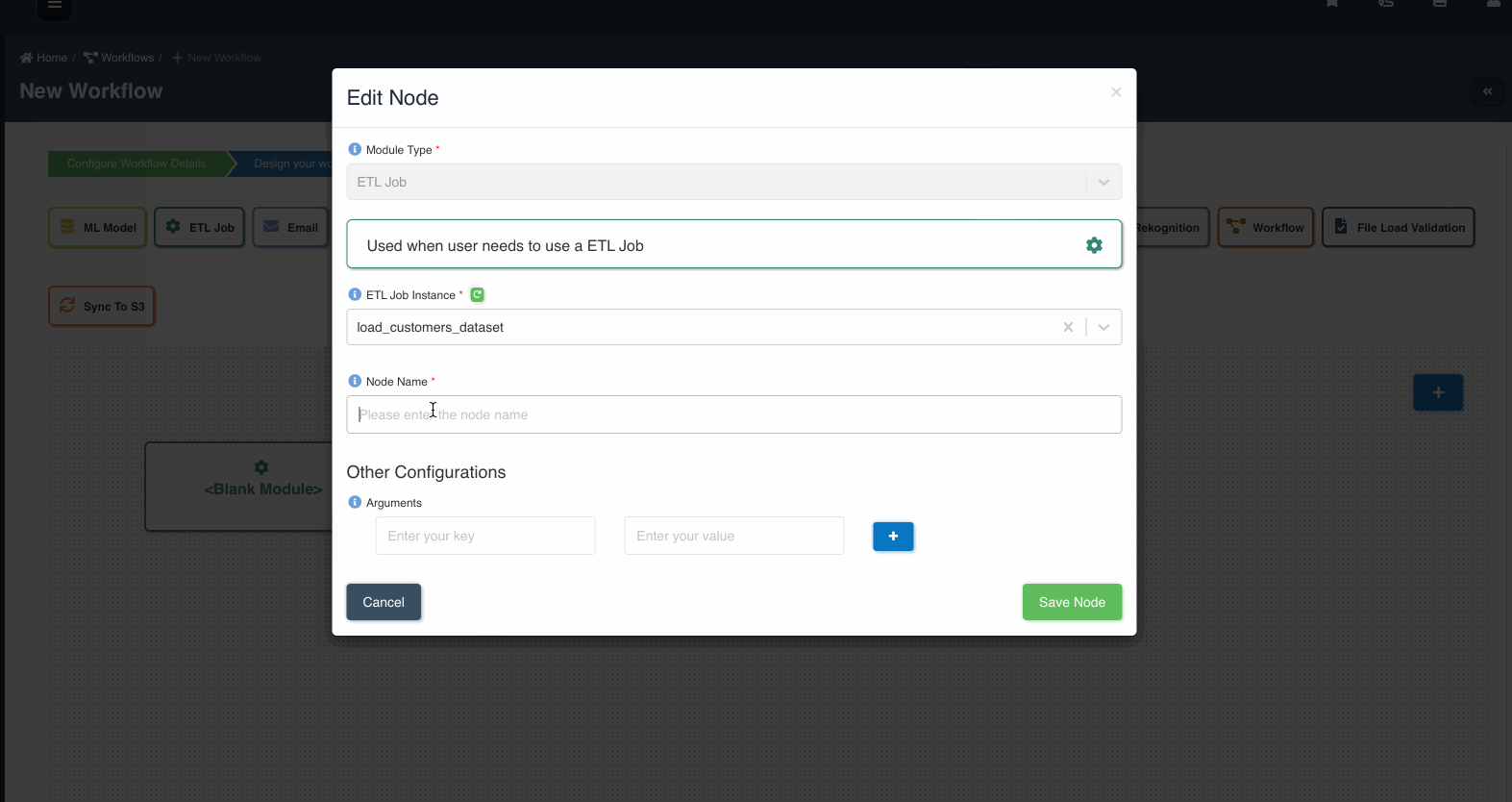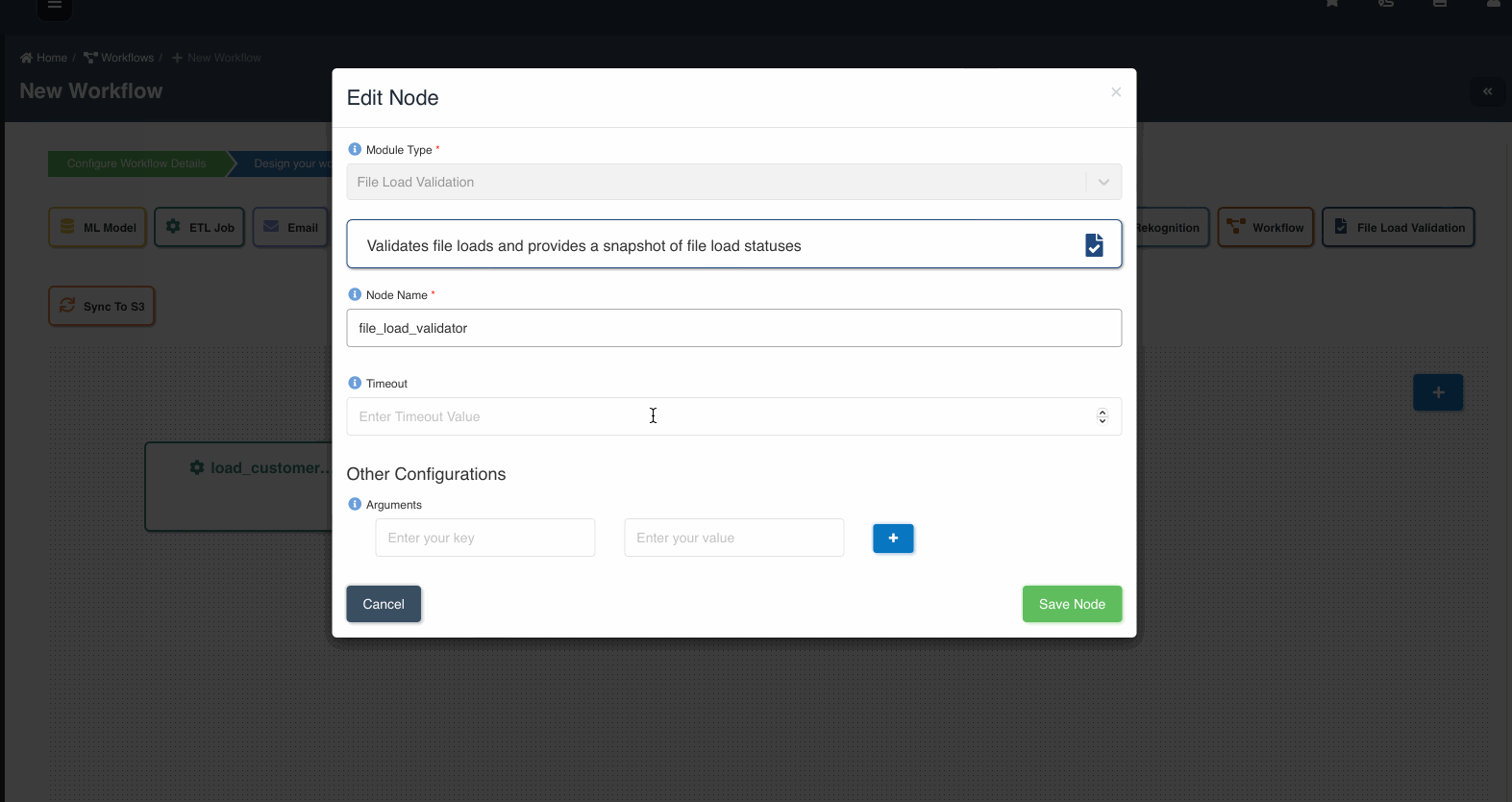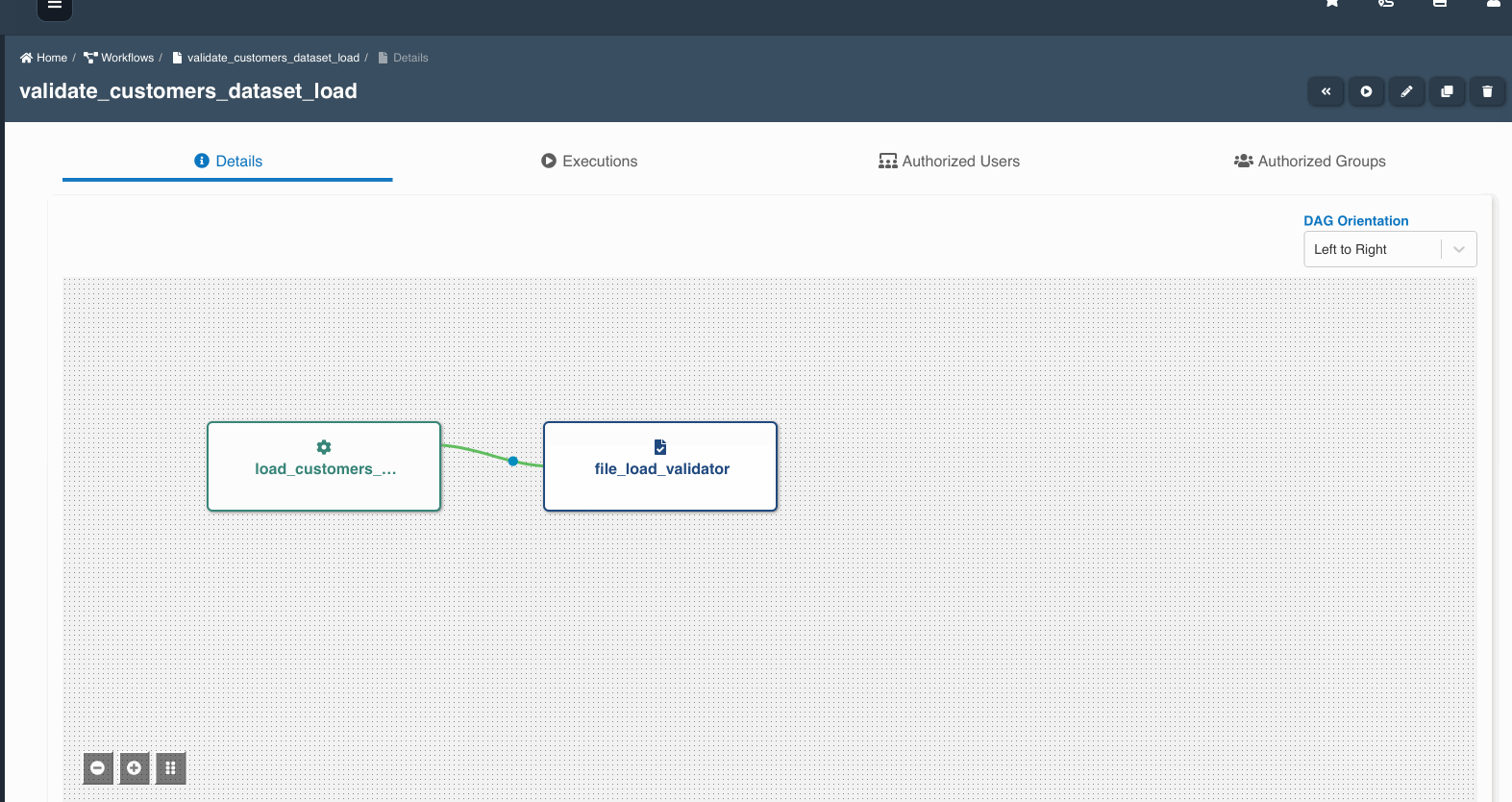
A file load validation node can only succeed an etl job, it cannot exist by itself or succeed other type of nodes in a workflow.
A sample workflow execution with file load validation looks as shown below:
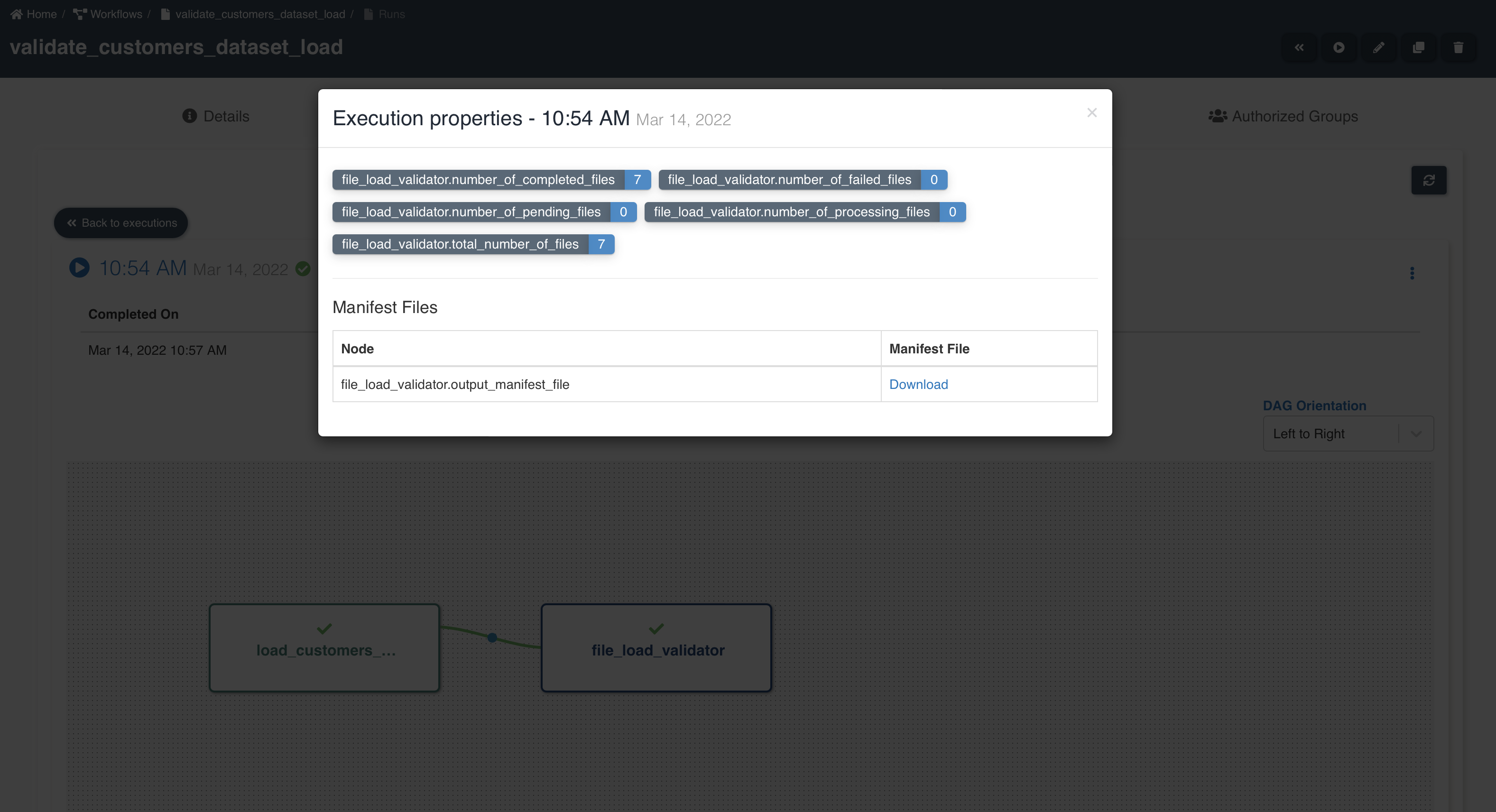
node_name.total_number_of_files - Number of files written by preceding etl job node.
node_name.number_of_completed_files - Number of files successfully validated and ready to be consumed from datasets.
node_name.number_of_processing_files - Number of files in process of being validated.
node_name.number_of_pending_files - Number of files in pending state waiting for success file to proceed further in processing. This is only applicable for datasets of type reload.
node_name.number_of_failed_files - Number of files that failed to be validated and are not available for consumption.
node_name.output_manifest_file - Manifest file showing list of file details.
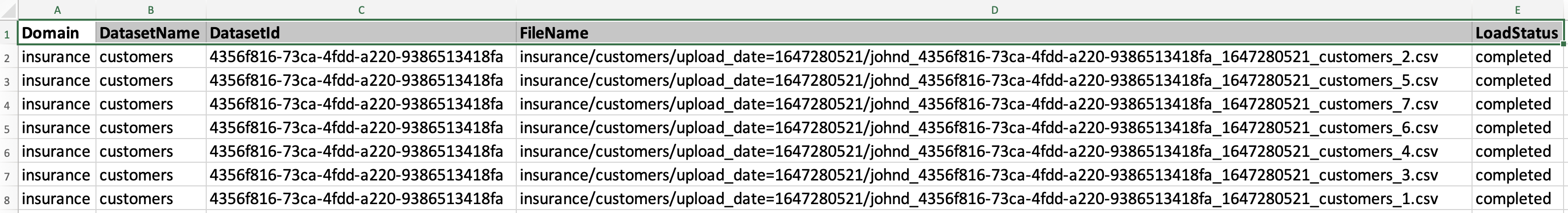
By clicking 'Download' button next to output manifest file, you will be able to download output manifest file as shown below:
It is advised to set number of concurrent runs to 1 on etl job when file load validation node is used. For advanced use cases which involve concurrent workflow executions, please refer to: Advanced usage of file load validation node
Sync To S3 Node
Sync To S3 operation for a dataset can now be executed from Connections, Schedules as well as Workflows in Amorphic console. Hence Sync To S3 operation for a specific dataset(under same domain) can only be executed from one of the Amorphic components at a time. If Sync To S3 is executed in parallel for a specific dataset from multiple places, the sync will succeed only for one execution and rest of the executions will fail with proper error message. This is done to avoid data inconsistency.
| Attribute | Description |
|---|---|
| Module Type | This field is preselected as 'Sync To S3' for file load validation nodes. |
| Node Name | Name given to the node for quick and easy identification. |
| Concurrency Factor | The number of datasets that user want to sync in parallel. Valid values range from 1-10. |
| Domain | The name of the domain for which datasets need to be synced from the Data Warehouse(Redshift or AuroraMySql) to S3 |
| Sync All Datasets | Indicates whether you want all datasets(Only registered datasets with TargetLocation as Redshift or AuroraMySql) in the domain to be synced(When set to 'Yes') or not. |
| Select Datasets | List of datasets(Only registered datasets with TargetLocation as Redshift or AuroraMySql) that you want to sync from the Data Warehouse to S3. This attribute is required only if you select 'No' for the 'Sync All Datasets' attribute. |
| Timeout | Optional field to set the timeout value (in minutes) for the node to stop execution. Default is set to 60 minutes. |
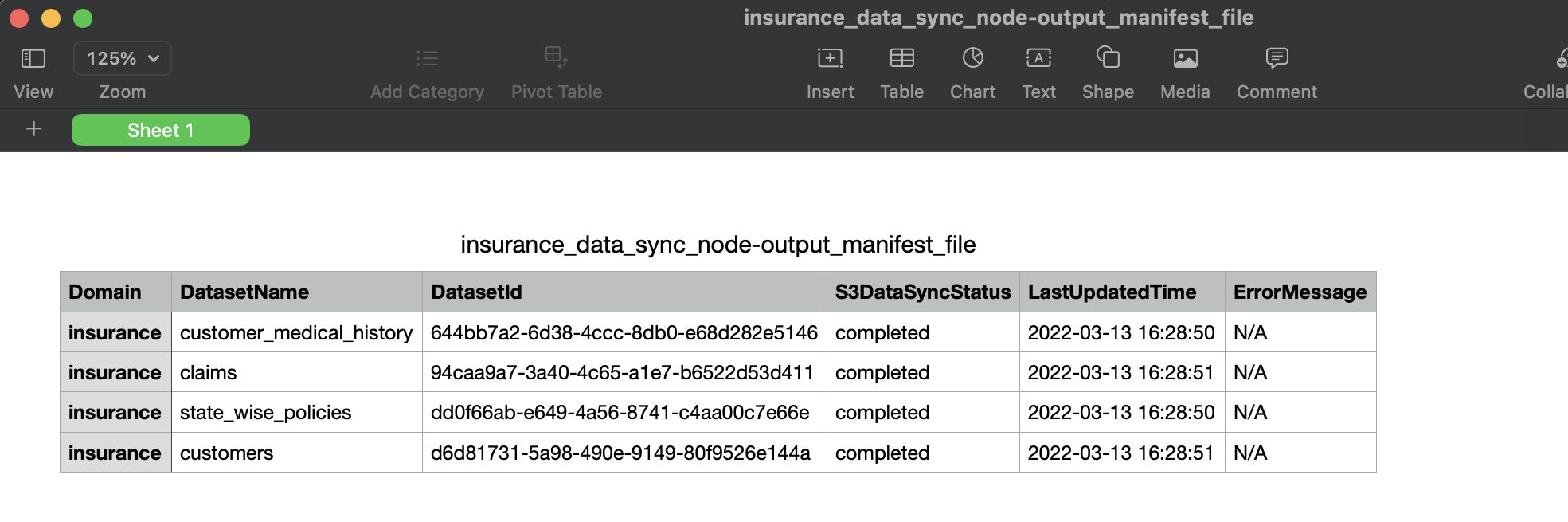
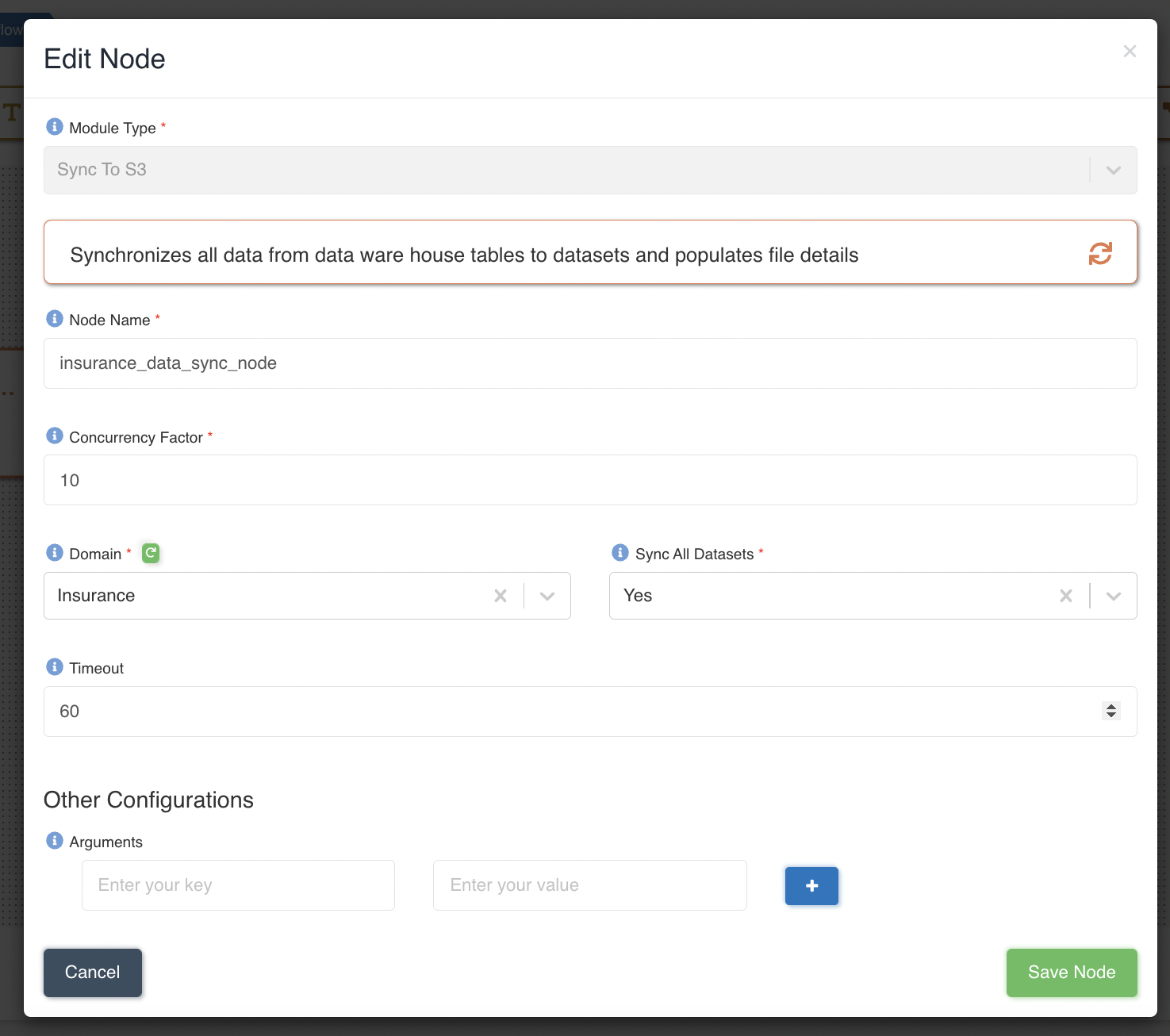
The following graphic shows how a Sync To S3 node can be created to sync data for all the datasets present in 'Insurance' domain & and at a time maximum 10 datasets will be synced in parallel:
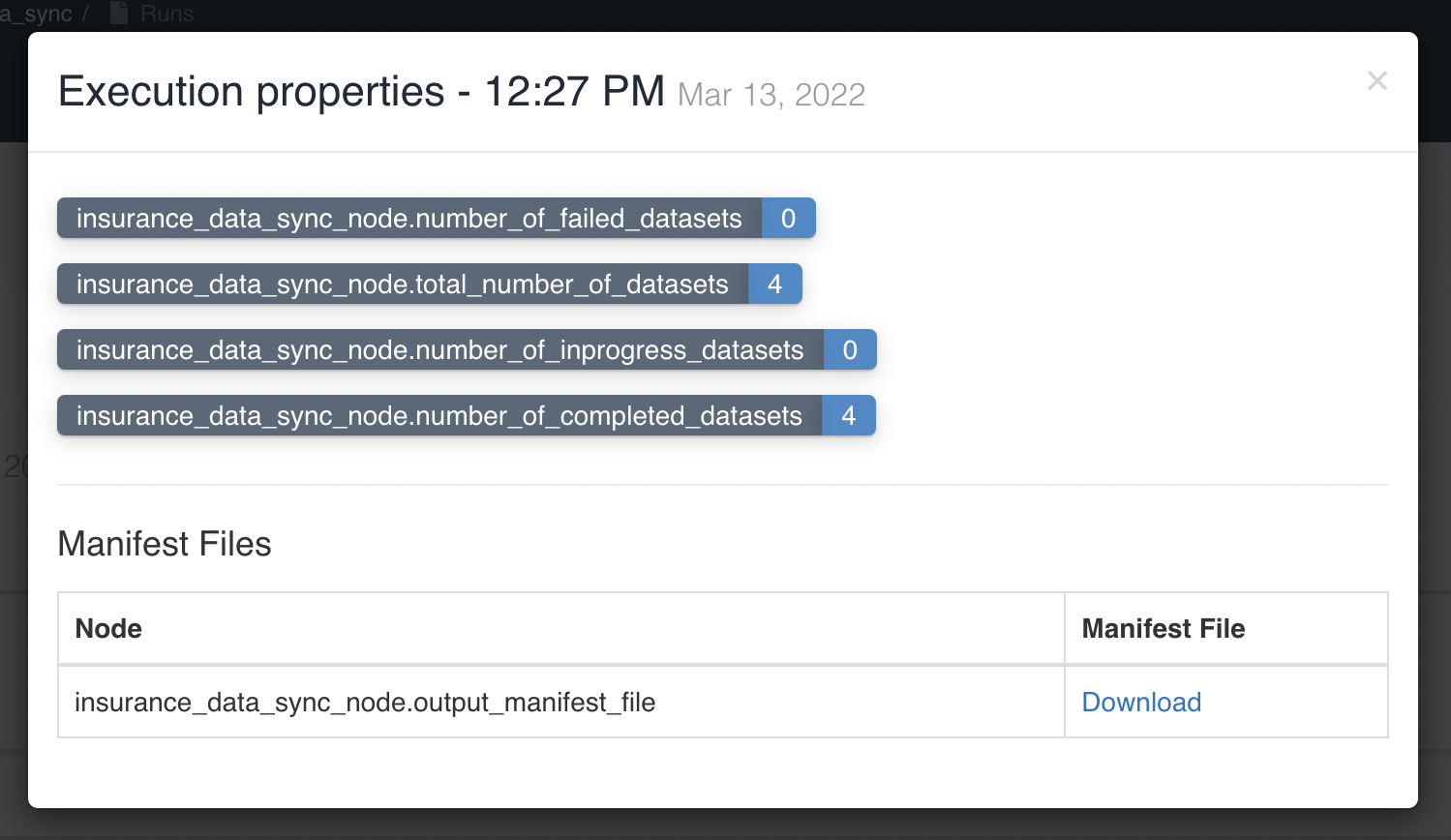
User can view/download the progress report of sync for all selected datasets in node execution properties. At a time, progress is updated only when a specific batch(based on Concurrency Factor, here 10) of datasets syncing is completed/failed. Below graphic shows execution properties of Sync to S3 Node:
User is given an option to click & 'Download' the manifest file in execution properties of workflow Sync To S3 node. Below graphic shows manifest file that is generated/downloaded after a workflow with sync to s3 node is executed: